This web page was produced as an assignment for an undergraduate course at Davidson College.
A single amino acid change in the spike protein may increase the transmissibility of SARS-CoV-2; However, more research is needed.
The COVID-19 pandemic thus far has infected 109,997,288 people, resulting in 2,435,145 deaths, according to the World Health Organization. Understanding the factors leading to the spread of the SARS-CoV-2 virus causing the pandemic is crucial to helping end this tragedy.
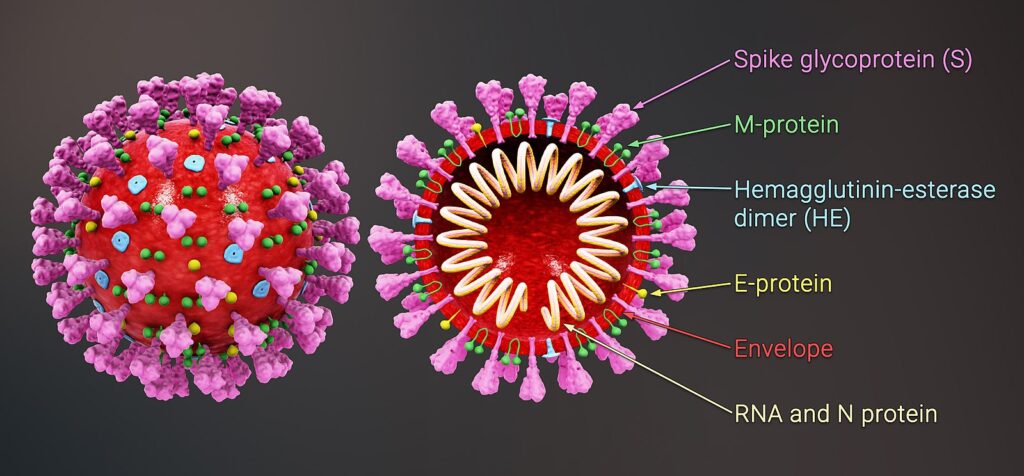
As the SARS-CoV-2 virus transmits, it accrues genomic mutations from errors in replication. While the vast majority of these mutations are harmful to the reproductive success of the virus, quickly being eliminated from the gene pool, occasionally a beneficial mutation will arise. Evolutionary theory then predicts that these beneficial mutations will then be selected for over time. In the case of SARS-CoV-2, a set of mutations resulting in the replacement of aspartic acid with glycine at position 614 of the virus’s spike protein has received considerable attention for its possible evolutionary benefit.
In laboratory tests, this D614G mutation has been shown to increase infectivity (Korber et al. 2020, Plante et al. 2020). Additionally, structural analysis has shown that the mutation allows SARS-CoV-2 to bind more easily to human cell surface receptors (Yurkovetskiy et al. 2020). Most importantly, the version of the virus with the mutated spike protein has overtaken the non-mutated version to become the dominant strain (Korber et al. 2020). However, all of these factors could have no real effect on the transmissibility of the virus, with its prevalence only the result of genetic drift (Korber et al. 2020).
Volz et al. analyzed 21,231 614G and 5,755 614D whole genome sequences sampled from different infections within the United Kingdom collected between January 29 and June 16, 2020, identifying phylogenetic clusters to determine if the 614G mutation conferred increased fitness. Despite the fact that most 614G clusters tended to be detected later in the pandemic, they were 59% larger than 614D clusters on average. As a direct result of the fact that this strain spread more, the 614G mutation soon became nearly omnipresent. However, this is not necessarily a result of reproductive benefit, Volz et al.‘s models showed that even if the mutation was not selected for, based on its previous trajectory, the prevalence of the mutation would increase in the population.
Fortunately, Volz et al. found there to be no difference in severity between the two studied strains of SARS-CoV-2. Interestingly though, however, younger patients were more likely to carry the strain with the mutated spike protein. The median age carriers of the 614D strain was 5 years older among females and 4 years older among males. This is possibly due to an increased viral load in younger patients resulting from 614G variants leading to higher detection rates. Through PCR testing, Volz et al. were able to determine that the 614G mutation was associated with a higher viral load.
Further complicating matters is the fact that there are multiple instances of the D614G mutation as well as reversions back to 614D. The existence of reversions implies that the 614D variant is still relatively fit within individual hosts. Additionally, there are a number of other variants that have occurred, with relative fitness levels yet to be determined. Furthermore, there are other mutations that have occurred elsewhere in the genome, potentially confounding any effects on fitness.
Given the wide variety of factors that influence the transmission of a virus in the real world, it is understandably difficult to confidently draw conclusions from an observational study. Furthermore, the ever evolving nature of the SARS-CoV-2 genome complicated efforts to understand it. Further research is obviously needed to conclusively establish that the D614G spike protein mutation leads to increased spread of COVID-19. This study only focused on the UK, hopefully by studying more sets of genomic data from different populations, we can continue to see similar patterns in terms of how this variant established its dominance.
Changes in the transmissibility of a circulating virus could have a major effect on the effectiveness of the pandemic response. In the time between the research on this study was done and it was published, a number of other SARS-CoV-2 variants have arisen and achieved widespread transmission. These variants are possibly even more transmissible than the ones discussed in this paper. Our knowledge of the SARS-CoV-2 genome must increase faster than the virus can mutate in order to be able to predict the future, not just explain the past. In fact, understanding the genomic evolution of SARS-CoV-2 is of even greater importance now than ever before in order to ensure that the vaccines that have been developed continue to be effective and save lives.
Stephen Skrynecki is a Sophomore Biology and Economics Double Major at Davidson College. Contact him at stskrynecki@davidson.edu.
Resources
Korber, B., W.M. Fischer, S. Gnanakaran, H. Yoon, J. Theiler, et al. Tracking changes in SARS-CoV-2 spike: evidence that D614G Increases infectivity of the COVID-19 virus. Cell. 2020;182(4):812-827.e19.
Plante, J.A., Y. Liu, J. Liu, H. Xia, B.A. Johnson,et al. Spike mutation D614G alters SARS-CoV-2 fitness. Nature 2020.
Volz, E., V. Hill, J. T. McCrone, A. Price, D. Jorgensen, et al. Evaluating the effects of SARS-CoV-2 spike mutation D614G on transmissibility and pathogenicity. Cell. 2021;184(1):64-75.e11.
Yurkovetskiy, L., X. Wang, K. E. Pascal, C. Tomkins-Tinch, T. Nyalile, et al. SARS-CoV-2 spike protein variant D614G increases infectivity and retains sensitivity to antibodies that target the receptor binding domain. Preprint. bioRxiv. 2020;2020.07.04.187757.
© Copyright 2020 Department of Biology, Davidson College, Davidson, NC 28036