This web page was produced as an assignment for an undergraduate course at Davidson College.
A multi-technique study of tumor cells reveals that DNA found off of the chromosome is extremely accessible by transcription factors, providing a strong clue for the progression of cancers.
In the year 2020, in the United States, approximately 606,000 people were projected to die as a result of cancer (Siegel et al. 2020). Cancer affects millions of people worldwide each year, and because of this, there is extensive research on numerous specifics that pertain to this disease. A very important topic of study is the role of oncogenes in the development and progression of cancers. In simple terms, oncogenes are genes that have the potential to cause cancer. Their precursors are proto-oncogenes and once these precursors become mutated, cancer-causing oncogenes may start to be expressed at high levels (Varmus et al 2005). It is somewhat unknown how these oncogenes become so highly expressed in cancer cells. To investigate this, researchers have begun to look past the information that the DNA sequences contain, and have focused on the information that can be obtained from the shape of DNA.
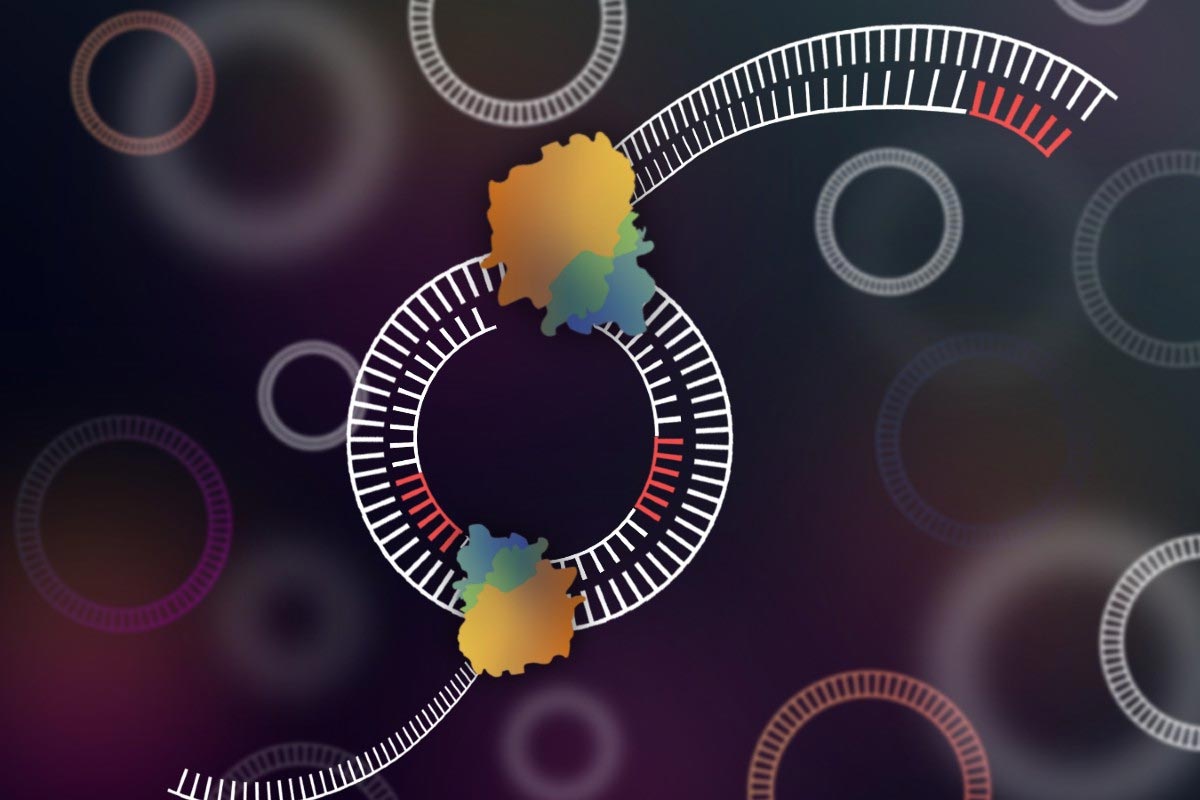
In human cells, the majority of the genetic information is found on chromosomes. It is important to note that chromosomes are made up of tightly wound chromatin. This compacted structure of the DNA on chromosomes makes it so that the transcriptional elements are confined to specific areas. While much of the human genome can be accounted for on the chromosomes, DNA can also be found off of the chromosomes in cells. This kind of DNA is more commonly referred to as extrachromosomal DNA (ecDNA). It is very likely that this ecDNA is not as compacted as chromosomal DNA, bringing into question certain transcription elements are able to affect these sequences. Researchers looked to see determine the regulatory effects that ecDNA has on cancers.
First, researchers needed to identify the shape in which ecDNA is configured to get a better understanding of the accessibility of the genetic material. To do this, a whole-genome sequence was analyzed to determine if amplified regions were linear or circular in glioblastoma cells. The amplified ecDNA in the tumor cells was confirmed to be circular and supported by other sequencing methods. Also, researchers confirmed that the amplified regions on chromosomes were linear. The new understanding of the circular shape of ecDNA in cancer cells provided an important base to study the expression patterns of the genes.
It has been previously identified that in cancer genomes, oncogenes are some of the most highly expressed genes. Researchers then wanted to investigate the effect that the circular shape of DNA has on transcription. Using an approach similar to the one that revealed the circular shape of ecDNA, researchers looked to see whether specific genes were amplified on circular ecDNA. They found that oncogenes that were found on ecDNA had significantly increased numbers of transcripts compared to the same genes that were not amplified with circularization. Oncogenes on amplified ecDNA showed higher copy numbers that the same genes amplified on linear structures. This information prompted new questions about the chromatin organization of ecDNA and the structural features that might contribute to the increased expression of oncogenes on ecDNA.
As noted earlier, much of the genetic material of the human genome is wound tightly around proteins and then further packaged. This makes much of the DNA inaccessible by transcription machinery making most of the human genome untranscribable in cells. This phenomenon is not seen in circular DNA. To determine the chromatin arrangement in ecDNA, researchers used a variety of techniques in conjunction with each other. With numerous different types of sequencing and assays, researchers saw that ecDNA was in fact packaged into chromatin and consisted of nucleosome units. However, these nucleosome units of ecDNA appeared to be much less compacted than their chromosomal counterparts. This reduction in compaction makes it much easier for transcription factors to interact with the genetic material. This is in line with the high transcription levels of oncogenes that are seen in cancer cells.
We see in this study that the shape of DNA in cancer cells have sizable effects on the progression of cancers. Specifically, shape and resulting accessibility of chromatin in extrachromosomal DNA makes it so that oncogenes in cancer cells are expressed at very high levels. Bacteria arranging their genetic material circular plasmids has been shown to be a powerful tool in gaining selective advantages over time (Delaney et al. 2018). This is a very similar concept that is occurring in these cancer cells. With more genetic material being organized and amplified off of the chromosome, cells in which this is occurring are more and more likely of oncogenesis, healthy cells being transformed into cancer cells. The next area of research is to find a way to work against the circular DNA. As previously mentioned, cancer impacts millions of individuals all across the world each year. It may be necessary of researchers to decrease the affinity of ecDNA for the transcription machinery. Of course, this technique will have to be very specific as it targets DNA that is a small minority of the entire genome. It is certainly a tall task, but the benefits will definitely reach far and wide.
Miles Davis is a junior biology major at Davidson College. Contact him at: midavis@davidson.edu
References
1. Siegel, R. L., Miller, K. D. & Jemal, A. Cancer statistics, 2020. CA. Cancer J. Clin. 70, 7–30 (2020). DOI: 10.3322/caac.21590
2. Varmus, H., Pao*, W., Politi, K., Podsypanina, K. & Du, Y.-C. N. Oncogenes Come of Age. Cold Spring Harb. Symp. Quant. Biol. 70, 1–9 (2005). DOI: 10.1101/sqb.2005.70.039
3. Delaney, S., Murphy, R. & Walsh, F. A Comparison of Methods for the Extraction of Plasmids Capable of Conferring Antibiotic Resistance in a Human Pathogen From Complex Broiler Cecal Samples. Front. Microbiol. 9, (2018).4. Wu, S. et al. Circular ecDNA promotes accessible chromatin and high oncogene expression. Nature 575, 699–703 (2019). DOI: 10.3389/fmicb.2018.01731
O’Neill, Mike. “New Tool Developed to Sequence Circular DNA.” SciTechDaily, 16 May 2020, scitechdaily.com/new-tool-developed-to-sequence-circular-dna/.
4. Wu, S. et al. Circular ecDNA promotes accessible chromatin and high oncogene expression. Nature 575, 699–703 (2019). DOI: 10.1038/s41586-019-1763-5
© Copyright 2020 Department of Biology, Davidson College, Davidson, NC 28036.