This web page was produced as an assignment for an undergraduate course at Davidson College
epigenetic profiling of circulating nucleosomes demonstrates potential ability as cancer diagnostic tool
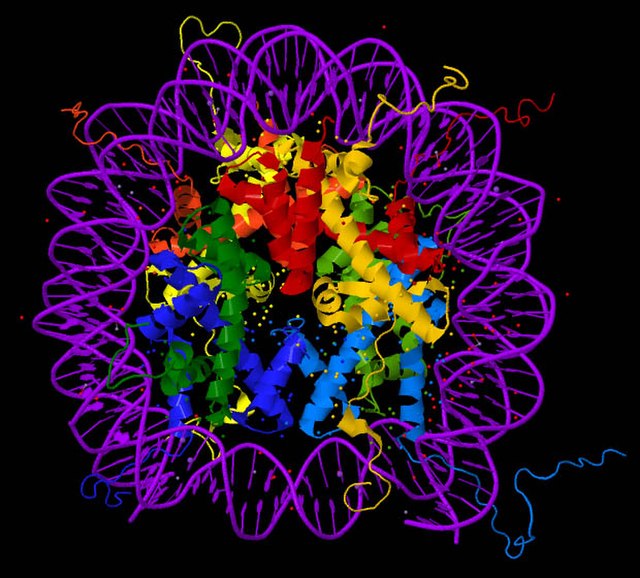
Cancer is the second leading cause of death in the US as of 2019 (CDC, 2019) and despite having significant improvements in diagnostic and screening strategies, the survival rate for advanced cancers detected in the later stages are still low (American Cancer Society, 2020). This can be improved with early diagnosis coupled with effective treatment since the cancer is less likely to have metastasized the sooner it is detected. Alteration of epigenetic modifications plays an important role in human cancer, for example, colorectal cancer (CRC) results from an accumulation of genetic and epigenetic alterations in colonic epithelial cells. Epigenetic biomarkers—which serve as a measurement of epigenetic alterations in tissues or internal fluids for the detection of diseases—are quickly emerging as a diagnostic tool for detecting cancer (Portela & Esteller, 2010). Nucleosomes are DNA wrapped around an octameric histone core protein to form the basic repeating unit of chromatin and the increased presence of nucleosomes in blood is suggestive of cancer (Van den Ackerveken et al., 2021). Currently, there are limited studies analyzing circulating nucleosomes or histones and the tools needed to investigate these potential epigenetic biomarkers directly from patient’s blood is also limited. Therefore, the authors of this study present a new method for the detection and the quantification of circulating nucleosomes and their associated histone epigenetic modifications in plasma samples. This new technique, Nu.Q Capture-MS, has the potential for discovering biomarkers for cancer diagnosis through the quantification of histone post-translation modifications (PTMs).
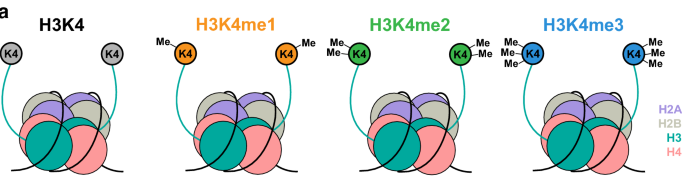
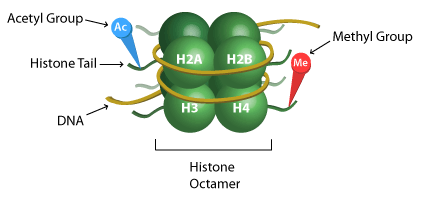
Epigenetic regulation can be complex, but one such mechanism of importance is the PTM of histones. Core histones (H2A, H2B, H3 and H4) are subject to PTMs which could ultimately modify the chromatin structure and lead to gene mis-expression. Epigenetic changes to the core histones are written in order of the histone, followed by the amino acid residue and epigenetic change, if any. For example, H3K27me4 is read as the tri-methylation of lysine 27 on histone 3. This can be confusing, so figure 1 is meant to serve as a guide for reading epigenetic profiles. Methylation (me) is one of three histone modifications mentioned in this study, the other two are acetylation (ac) and citrullination (cit). To perform proteomic analysis of PTMs present on circulating nucleosomes, the authors developed an enrichment method based on immunoprecipitation—a technique used to separate proteins that are bound to a specific antibody from the rest of a solution—followed by liquid chromatography. This technique for capturing nucleosomes coupled with mass spectrometry (Nu.Q Capture-MS) allows the epigenetic profiles analysis of circulating nucleosomes (Van den Ackerveken et al., 2021).
A pilot study utilizing this new protocol was conducted by using plasma samples from nine CRC patients and nine healthy donors. Among the histone peptides identified from circulating nucleosomes, 23 histone proteoforms were found to be differentially represented in plasma from CRC patients or healthy donors, supporting prior research of cancer patients having higher levels of nucleosomes in their blood (Rahier et al., 2017). Further analysis of these histone peptides led to the characterization of 13 distinct histone PTMs. Levels of H3K9 methylation (-Me1, -Me2, -Me3), H3K27 methylation (-Me1 and -Me2), H3 acetylation (located at K14, K18, K23, K27, and K36) as well as levels of H2A1R3 citrulline (H2A1R3Cit), were significantly increased in CRC compared to healthy controls. To identify the fraction of tumor-derived nucleosomes in blood from cancer patients, levels of histone PTMs present in plasma were compared to the levels present in tumor resection (CRC tissues) or normal adjacent tissues (NAT) from the same patients. Tissue analysis revealed numerous histone peptides differentially abundant in CRC tissue vs NAT and that several of these peptides exhibited distinctive histone PTMs. To determine which of the histone PTMs were more likely to be tumor-derived as opposed to tumor-associated, the authors compared the set of histone PTMs identified in CRC tissues to that of histone PTMs peptides significantly mis-regulated in the plasma of CRC patients. Three common histone PTMs were found to be similarly dysregulated in plasma and tumor tissue from the same patients (H3K27Me2, H3K27Ac, and H2A1R3Cit) suggesting that the presence of these histone PTMs on circulating nucleosomes could be tumor-derived and reflect the epigenetic signature of the tumor.
The Nu.Q Capture-MS protocol demonstrated that the epigenetic pattern of circulating nucleosomes can be analyzed by mass spectrometry and that the epigenetic profile of circulating nucleosomes is different between CRC patients and healthy donors. This newly developed technique has significant implications for discovering epigenetic biomarkers of diseases and as a tool for diagnostic purposes. Cancer is an umbrella for an immense number of diseases, with many still deserving more research for improvements in detection and treatment. This novel approach to epigenetic profiling of circulating nucleosomes presented here has the potential to tremendously impact the prognosis of cancer, generally, by detecting and diagnosing the disease in its early stages. This will still need to be coupled with effective treatment, but early detection is an essential step towards decreasing cancer-related deaths.
Dreylan Hines is an undergraduate student at Davidson College pursing a Bachelor’s degree in Biology. Contact him at drhines@davidson.edu.
© Copyright 2020 Department of Biology, Davidson College, Davidson, NC 28036.
Access the home page here.
REFERENCES
Colorectal Cancer Stages: Rectal Cancer Staging: Colon Cancer Staging. (2020, June 20). Retrieved from https://www.cancer.org/cancer/colon-rectal-cancer/detection-diagnosis-staging/staged.html
Leading Causes of Death. (2021, March 01). Retrieved from https://www.cdc.gov/nchs/fastats/leading-causes-of-death.htm
Portela, A., & Esteller, M. (2010). Epigenetic modifications and human disease. Nature biotechnology, 28(10), 1057–1068. https://doi.org/10.1038/nbt.1685
Rahier, J. F., Druez, A., Faugeras, L., Martinet, J. P., Géhénot, M., Josseaux, E., Herzog, M., Micallef, J., George, F., Delos, M., De Ronde, T., Badaoui, A., & D’Hondt, L. (2017). Circulating nucleosomes as new blood-based biomarkers for detection of colorectal cancer. Clinical epigenetics, 9, 53. https://doi.org/10.1186/s13148-017-0351-5
Van den Ackerveken, P., Lobbens, A., Turatsinze, J. V., Solis-Mezarino, V., Völker-Albert, M., Imhof, A., & Herzog, M. (2021). A novel proteomics approach to epigenetic profiling of circulating nucleosomes. Scientific reports, 11(1), 7256. https://doi.org/10.1038/s41598-021-86630-3